Terry Nguyen and Justin suggested that I write about Computational Biology. So here it is :-)
Whenever someone asks me what is my major, I'll say that I am part Computer Science and part Computational Biology since my undergraduate was in Computer Science while my HYP and current work is in Computational Biology. I have to confess that I'm not a real biologist and my biology knowledge is fairly limited, but I have a healthy curiosity in new developments in biology. The diversity and complexity of life has always fascinated me.
Computational biology is the application of computational techniques for tackling problems in biology. The problems can be broadly classified into two types. The first is to assist in the design/interpretation/analysis of high throughput experiments, such as micro arrays, tandem mass spectroscopy and DNA sequencing machines. The second is to discover meaningful patterns from biological data to improve our understanding of the underlying biological mechanisms.
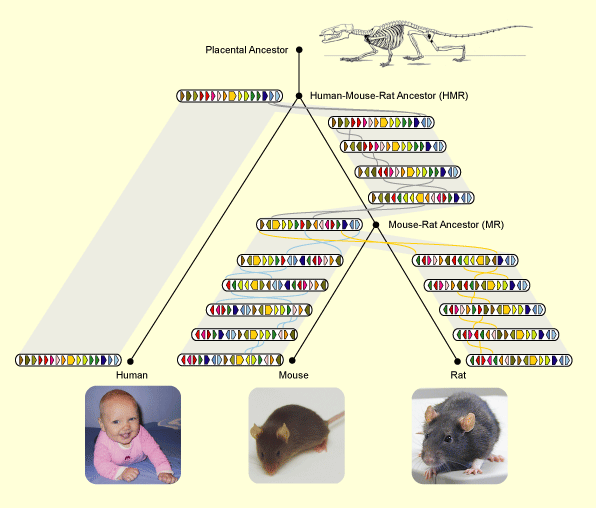
My own work in the comparative genomics of gene order is from the second category. The data I am considering is a set of gene orders, where a gene order is the sequence of genes that makes up the genome of a species. Although many species share a large number of genes, the arrangement of these genes are different due to large scale rearrangement events (see Figure 1 above) that has the effect of shuffling the genes around.